Chapter 4 Taxonomy Profiling
Taxonomic profiling refers to the annotation and classification of metagenomics sequencing reads with taxonomic information. Due to the high complexity of metagenomics data, long-read shotgun sequencing offers many advantages over short-read sequencing for taxonomic classification. In this module, we will get a taste of long-read metagenomics sequencing data, and learn to classify and visualize taxonomy in Galaxy platform using tools like Kraken 2 to annotate and Krona plot to visualize taxonomy.
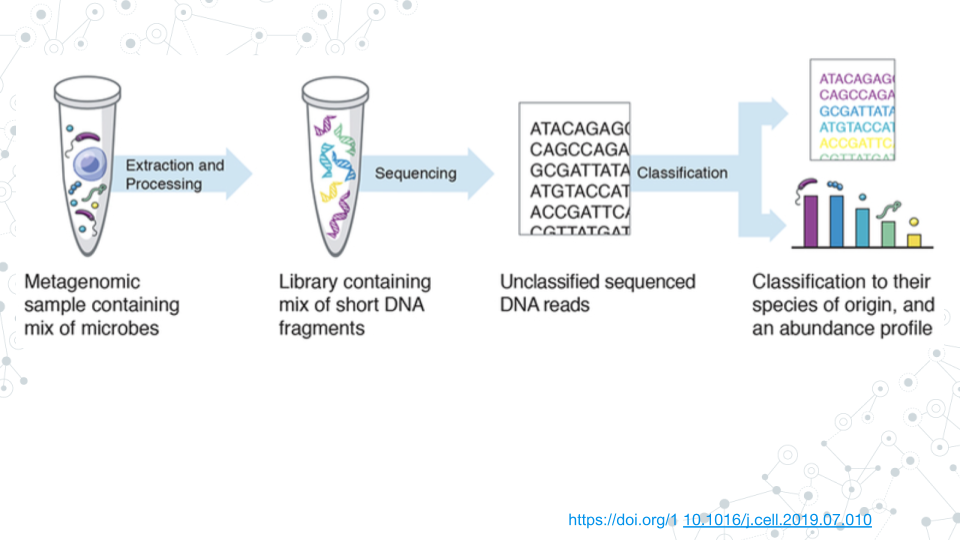
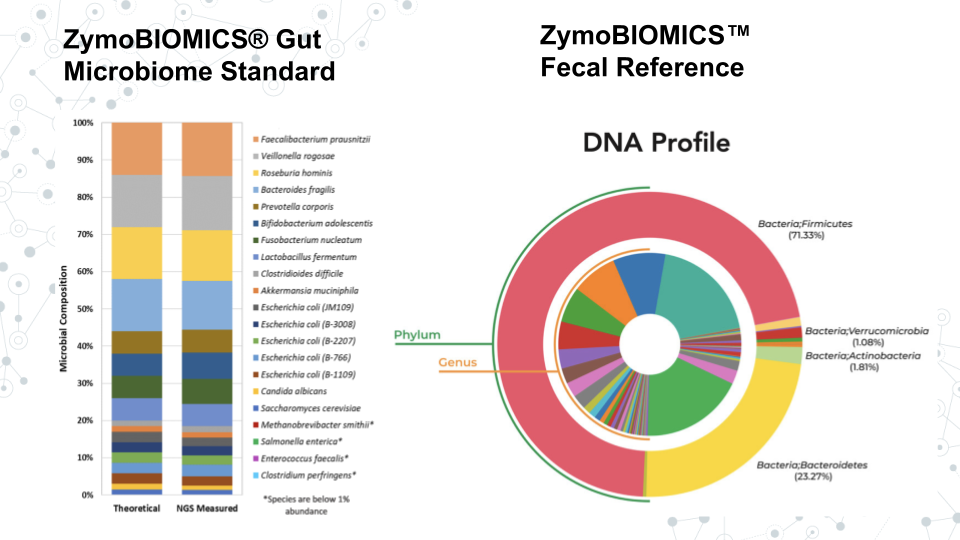
This Taxonomy Profiling module offers hands-on experience with real data. We will use lecture material to learn about taxa and related concepts like taxonomic classification and species abundance. In the Activity portion of the module we will explore taxonomy of gut microbiome standard from Zymo Research using output of Kraken 2. This standard is a sample of known microbes with expected relative abundances. Through this activity we will become familiar with the structure of our taxonomy data and do some basic analyses. In Prelab activity, we will have a chance to perform taxonomy profiling ourselves using raw sequences from the gut microbiome standard on Galaxy platform to reproduce the results we analyzed in the previous activity, using the tools Kraken 2 (for assignment of taxonomy to sequencing reads) and Krona pie charts (for visualization). Subsequently, we will do a Project activity, where we will taxonomically classify a soil metagenome sample which serves as an example of an uncurated novel data. We will compare our results with the gut microbiome standard over a variety of metrics to learn what to expect when handling real data.