
Chapter 3 Microbial Genomes
Microbial genomes range in size and organization. Despite their small size and having fewer number of coding genes, bacteria are more taxonomically diverse, more abundant and more successful in colonizing neew habitats - they are present in virtually every environment on earth!. This is in part due to bacteria having very ‘busy’ genomes with a high proportion of protein-coding functional genes and regions (and very little noncoding DNA and introns) which provide a source of innovation, novelty and adaptation e.g. by simply altering (mutating) functional genes (P.C. Kirchberger et al, 2020). In contrast, eukaryotic genomes mostly consist of noncoding DNA, and a lot of mutations are bound to occur in those abundant noncoding regions.
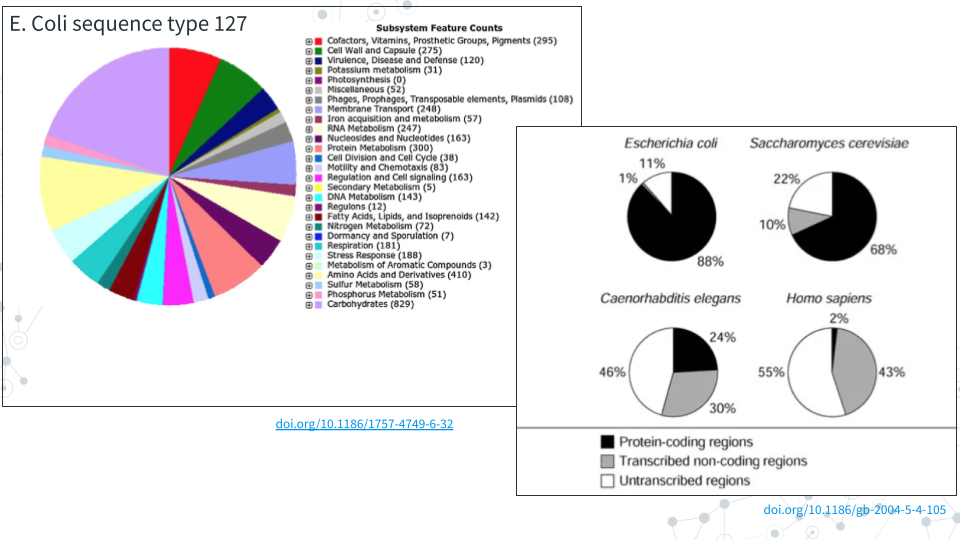
In this Microbial Genomes module, we will learn about microbial genomes and microbial databases. In the Pre-lab activity, we will use E. coli to explore NIH genetic sequence database GenBank, bacterial databases such as BV-BRC and tree of life database Lifemap. For Project activity, we will continue microbial genome exploration using a published metagenome-assembled genomes or MAGs, which are microbial genomes assembled (reconstructed from) metagenome sequencing data. We will learn about the taxonomy of the MAG, its size, and assembly statistics and about its genes.