Chapter 5 Finding AMRs
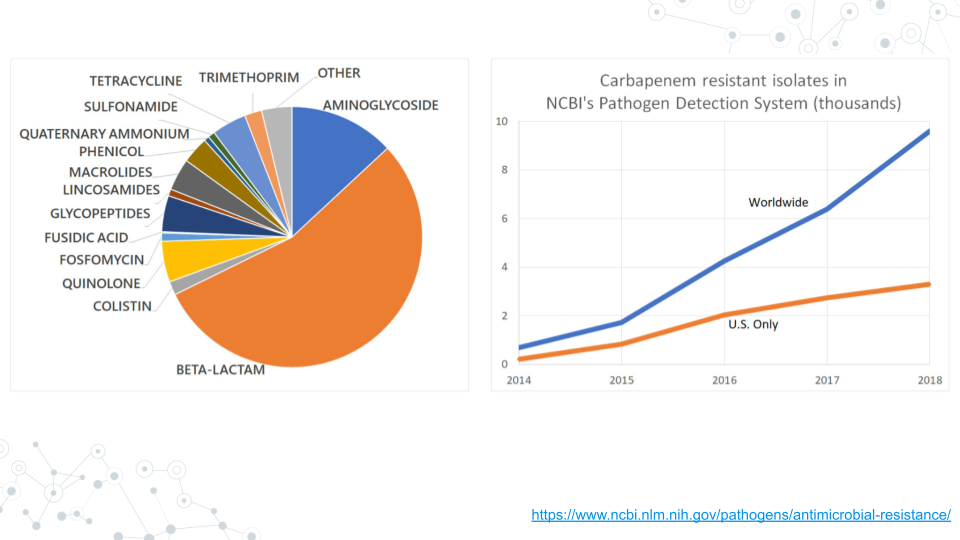
Antimicrobial resistance genes (AMR genes) allow microbes to counteract the effects of antimicrobial drugs used to treat infections. Databases such as the NCBI Pathogen Detection Reference Gene Catalog and the Comprehensive Antibiotic Resistance Database contain thousands of curated resistance genes and help make AMR-related data more widely available.
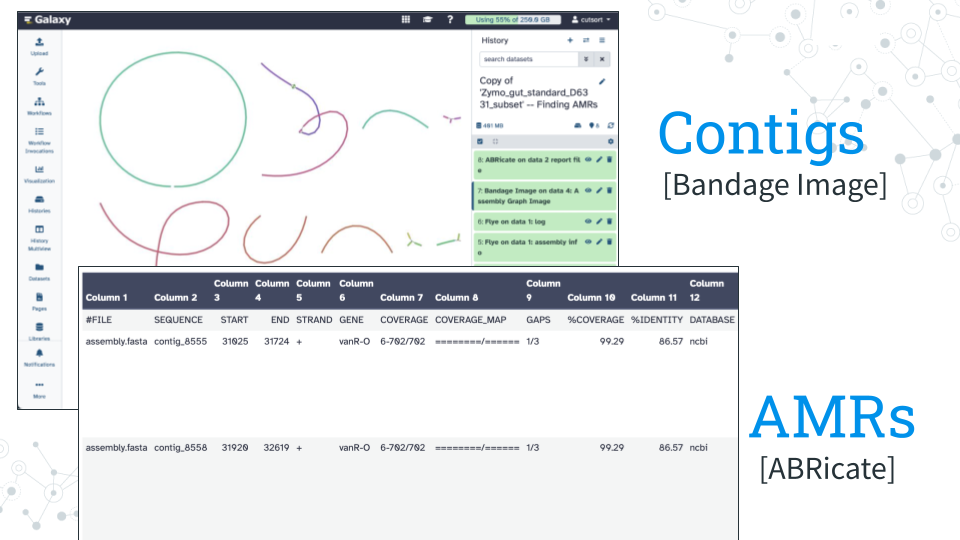
In the Finding AMRs module, the lecture material will cover antibiotic resistance and concepts of genome assembly and genome annotation. In the Prelab activity, using Galaxy platform, we will assemble the prototype of gut microbiome using gut microbiome standard from Zymo Research. We will first assemble the genomes into contigs, then visualize the contigs using Bandage Image tool, followed by screening contigs for AMRs using the tool ABRicateantimicrobial genes using a variety of databases including the NCBI database.
A similar strategy can be used to screen for virulence factors using databases such as the Virulence Factor Database (VFDB). For Project activity, students will perform soil genome assembly and in addition to annotating soil metagenomes with AMRs, we will assemble contigs into larger MAGs, and learn about annotating MAGs with tools like GTDB-Tk.